Lecture 2
Environments
Environments are special data structures that are responsible for handling the names of variables and for looking up variables when requested. You have been interacting with environments for as long as you have been using R. Whenever you create a variable in R, what you have actually done is created an object then created a slot in the global environment (the environment where user action in R generally takes place) that points to that object. If an object has no slot in an environment pointing to that object, R automatically destroys it during a process known as garbage collection.
The function ls() lists the objects contained in an environment. By default,
it lists what’s in the global environment.
## [1] "_Mean.1_" "%s%" "%s0%" "1.Mean_"
## [5] "Awesome sauce!" "collector" "error_obj" "f_num"
## [9] "i" "idx" "increment" "l"
## [13] "m" "n" "new_collector" "paste_"
## [17] "s_num" "stat" "sum" "t_num"
## [21] "u" "x" "z.stat"## [1] "_Mean.1_" "%s%" "%s0%" "1.Mean_"
## [5] "Awesome sauce!" "collector" "error_obj" "f_num"
## [9] "i" "idx" "increment" "l"
## [13] "m" "n" "new_collector" "paste_"
## [17] "s_num" "stat" "sum" "t_num"
## [21] "u" "x" "z.stat"## [1] "_Mean.1_" "%s%" "%s0%" "1.Mean_"
## [5] "Awesome sauce!" "collector" "error_obj" "f_num"
## [9] "i" "idx" "increment" "l"
## [13] "m" "n" "new_collector" "paste_"
## [17] "s_num" "stat" "sum" "t_num"
## [21] "u" "x" "z.stat"## [1] "_Mean.1_" "%s%" "%s0%" "1.Mean_"
## [5] "Awesome sauce!" "collector" "error_obj" "f_num"
## [9] "i" "idx" "increment" "l"
## [13] "m" "n" "new_collector" "paste_"
## [17] "s_num" "stat" "sum" "t_num"
## [21] "u" "x" "z.stat"The function environment() lists the current environment.
## <environment: R_GlobalEnv>In fact, we can access objects in environments similarly to how we access
objects in lists. However, environments are not just a different kind of list.
We’ll get into that in a second; let’s first see how environments have similar
syntax to lists. We can create a new environment with the function new.env().
Objects in the environment can be referenced using $, like so:
my_env <- new.env()
my_env$x <- 1
my_env[["y"]] <- 2
my_env[["awesome sauce"]] <- 3
my_env[["x"]] + my_env$y + my_env$`awesome sauce`## [1] 6## [1] "awesome sauce" "x" "y"However, despite these similarities, there are important distinctions between
environments and lists, such as objects saved in an environment are unordered;
there is no “first” item. So in the above code, my_env[1] and my_env[[1]]
make no sense. Names in environments are unique, and environments have reference
semantics. Additionally, you don’t remove objects from environments by setting
them to NULL, as you would with a list. Instead, you have to use the function
rm(), like so:
There are two fundamental ingredients needed for an environment: a frame
(which are the name-object bindings demonstrated above) and the environment’s
parent environment, which is an environment that “contains” the environment.
There is only one environment that does not have a parent: the empty
environment (created automatically and referenced with emptyenv()). Otherwise,
every environment has a parent. Aside from emptyenv(), two other important
environments are the global environment (referenced via globalenv() and
represents the environment that the user generally works in) and baseenv()
(the environment of the base package).
When we create an environment, we can declare its parent like so:
## <environment: 0x55910d17a400>## <environment: 0x55910d17a400>By default, the parent of an environment created via new.env() is the global
environment. When using environments as data structures (say, as a substitute
for a list), consider making the parent the empty environment.
Due to the parent-child nature of environments, we could say that any environment has a sequence of ancestors, consisting of the parent of a given environment, the parent of the parent environment, and so on. The only environment that does not have a parent is the empty environment. Furthermore, the empty environment is the ultimate ancestor of all environments.
To compare environments, we use a function called identical(); we cannot use
==. I demonstrate use of identical() below:
## <environment: package:stats>
## attr(,"name")
## [1] "package:stats"
## attr(,"path")
## [1] "/usr/lib/R/library/stats"## [1] FALSE## [1] FALSE## [1] TRUEFunctions and Environments
Why does this discussion matter? Users generally are not creating environments. Instead, environments are generated automatically when functions are called. Users don’t notice because those environments are often instantly destroyed when the function completes and terminates. However, this matters when understanding and creating closures.
Consider the following function:
## x is 30
## y is 20## [1] 10## [1] 20## x is 30
## y is 50## [1] 40## [1] 50What happened here? Here’s a step-by-step breakdown of the above code sequence:
- Variables
xandywere created in the global namespace with some initial values. - The function
f()was defined, then called. Whenf()was called, a new environment was temporarily created, with the global environment being its parent. - A variable
xwas defined in the environment wheref()operated. Thisxis distinct from thexthat was defined in the global environment, since they were defined in two separate environments. This is the reason why the value ofxin the global environment was not changed. - When
xwas referenced inf(), R first looked inside the active environment, the temporary one created when the function was called.xwas found there, so thexthat existed in the global environment was ignored. Whenywas referenced, noywas found in the active environment, so R looked in the parent environment, which was the global environment. A definition forywas found there, and so it was the variable referenced. - We later changed the value of
xandyin the global environment. The first temporary environment from whenf()was called the first time was destroyed when the function finished its execution. When we called the function a second time, a brand new environment was created, with the global environment as its parent.xwas still created in this new environment, with its same value as before. The change toxin the global environment didn’t affect the function’s execution. The reference toy, though, did cause different behavior, since theyreferred to in the function was theythat existed in the global environment.
What if we wanted to change the value of a variable in the global environment
from the function? We could do so by using <<-, which will only create a
variable if no name is found in any of the current environment’s ancestors, and
if no reference is found, the variable is created in the global environment.
<<- is demonstrated below:
## x is 30
## y is 20## [1] 30## [1] 20Now this time x was changed in the global environment. This is because when
<<- was called, it looked for x in the active environment, didn’t find it,
then looked in the parent of the active environment, which was the global
environment. It found x there, and modified its value. When x and y were
referenced in the function, both of them were from the global environment
(before, only y was a global variable).
Namespaces
A namespace is a special environment associated with a package. Each package has its own namespace that is attached to the global environment when the package is loaded. However, thanks to namespaces, we can even reference functions in packages without even loading the package.
We reference objects in namespaces via either :: or :::, using syntax such
as namespace::object or namespace:::object. The difference between :: and
::: is that :: only accesses public objects, or objects that the package
authors have marked as available to all of R when the package is referenced.
::: accesses all objects in a package, including private ones that the
package authors did not intend to make available to other code and probably
don’t want to be available. While referencing package objects via ::,
referencing via ::: is unsafe and should be avoided.
For example, the function mvrnorm() from the package MASS allows for
simulating random variables that are joinly Normal. We can use this function
without explicitly loading MASS via :: like so:
## [,1] [,2]
## [1,] 0.6140040 0.4498941
## [2,] 1.3633067 0.7191522
## [3,] 1.1062482 0.4535209
## [4,] -0.8262249 0.1702595
## [5,] 0.1940201 1.9191629
## [6,] 1.3518935 0.6685813
## [7,] 1.1313676 1.5529045
## [8,] 0.5231649 1.9132617
## [9,] -0.5563917 0.2959209
## [10,] 0.3515644 0.1049541Why do namespaces matter? Consider the function sd() which is a function from
the packages stats that computes standard deviations. Reading the function’s
code reveals that it calls the function var(), also from stats, to compute
the standard deviation.
## function (x, na.rm = FALSE)
## sqrt(var(if (is.vector(x) || is.factor(x)) x else as.double(x),
## na.rm = na.rm))
## <bytecode: 0x55910bedef50>
## <environment: namespace:stats>The following code reveals that both sd() and var() “live” in an environment
that is not the global environment.
## <environment: namespace:stats>## <environment: namespace:stats>What if we change var? Will doing so break sd()?
## [1] 1.058425## [1] 1.028798## [1] 10## [1] 1.028798The output of sd() is unchanged. This is because sd() refers to the version
of var() that lives in its namespace, and doesn’t care about the version that
lives in the global environment. The var() in sd()’s namespace can’t be
modified in an active R session; one would have to modify the code of the
stats package in order to change var(). Therefore, sd() performs as
expected.
If we wanted to go back to the old var(), we could do so via the following
code:
## function(x) {10}## function (x, y = NULL, na.rm = FALSE, use)
## {
## if (missing(use))
## use <- if (na.rm)
## "na.or.complete"
## else "everything"
## na.method <- pmatch(use, c("all.obs", "complete.obs", "pairwise.complete.obs",
## "everything", "na.or.complete"))
## if (is.na(na.method))
## stop("invalid 'use' argument")
## if (is.data.frame(x))
## x <- as.matrix(x)
## else stopifnot(is.atomic(x))
## if (is.data.frame(y))
## y <- as.matrix(y)
## else stopifnot(is.atomic(y))
## .Call(C_cov, x, y, na.method, FALSE)
## }
## <bytecode: 0x55910c0436d0>
## <environment: namespace:stats>Closures
The above discussions about environments may have been enlightening, but the primary motivation was to allow discussion of writing functions that can return closures, which are also functions (the term simply distinguishes a function created by another function from usual functions).
Here’s an example of a function that returns closures:
## [1] "function"## [1] 3 4 5## [1] 201 202 203Closure constructors can be powerful tools, but to the uninitiated they can be
mysterious. Why is it, for example, that both f() and g() remember the value
i was set to when they were created?
The answer is environments. When we see what environment the functions “live”
in, we discover it’s not the global environment. Instead, these functions
still use the “temporary” environments created when the function incrementer()
was invoked; these environments were actually not destroyed when the function
ended since other functions were created in those environments, then returned,
and thus those environments are still needed. We in fact see that the
environments the closures use are not the global environment:
## <environment: R_GlobalEnv>## <environment: 0x55910c470c90>## <environment: 0x55910c6e84a8>And in fact we see that the i values from those function calls live on in
those environments.
## [1] "i"## [1] 2## [1] 200These features give closures their true power. For example, we can create functions that remember how many times they were called.
elephant_func <- function() { # Cuz elephants never forget
calls <- 0
function() {
calls <<- calls + 1
calls
}
}
e1 <- elephant_func()
e1()## [1] 1## [1] 2## [1] 3## [1] 1## [1] 2## [1] 4This set of functions may be more difficult to unparse than the examples we saw
before, yet the only difference in the logic is that the parent environment of,
say, e1()’s execution environment is not the global environment anymore but
instead the environment created upon e1()’s birth. This is distinct from the
parent environment of any environment created by e2()’s execution.
Here’s one application of closures. Suppose that we want to examine multiple confidence intervals for any given data set and we want an easy interface for doing so. With closures we can create easy interfaces for changing the parameters of confidence intervals, perhaps changing confidence levels or making them one-sided or two-sided. The following code is a function factory implementing this idea:
dat_ci <- function(dat) {
function(C = 0.95, type = "two.sided") {
alternative <- switch(type,
"two.sided" = "two.sided",
"upper" = "less",
"lower" = "greater"
)
c(t.test(dat, alternative = alternative, conf.level = C)$conf.int)
}
}
x1 <- rnorm(10)
c1 <- dat_ci(x1)
c1()## [1] -0.6752207 0.5351377## [1] -0.5604414 0.4203585## [1] -0.9394474 0.7993645## [1] -Inf 0.6847582## [1] -0.2349545 1.1986683Often in statistics we want to treat a data set as fixed but allow for, say, a parameter, to vary. Closures make doing this easier. Let’s consider, for example, the sum of square errors:
\[SSE(\theta) = \sum_{i = 1}^n (x_i - \theta)^2\]
We want to find \(\theta\) that minimizes the sum of square errors; we might call such a \(\theta\) a best predictor for an observation, or a good statistic describing the location of \(x_i\). Calculus reveals that the value of \(\theta\) that minimizes \(SSE(\theta)\) is \(\theta = \bar{x}\), the sample mean. But let’s see if we can avoid calculus.
We can write a function that produces closures that compute \(SSE(\theta)\) for
input \(\theta\) given a fixed data set. We would like this function to produce a
vector of SSEs if given a vector of values of \(\theta\). We do this by actually
returning a vectorized version of the \(SSE(\theta)\) function using the
Vectorize() function (which is a functional that returns closures). The final
implementation is below:
sse_computer <- function(x) {
f <- function(t) {
if (!is.numeric(t) || length(t) > 1) stop("Invalid t")
sum((x - t)^2)
}
Vectorize(f)
}If we wished to plot \(SSE(\theta)\) for a given data set, we can do so via the
curve() function, which is a plotting function with an interface friendly to
mathematical functions (try curve(exp, -2, 2) if you want a quick demo; this
is \(e^x\) plotted from \(x = -10\) to \(x = 10\)).
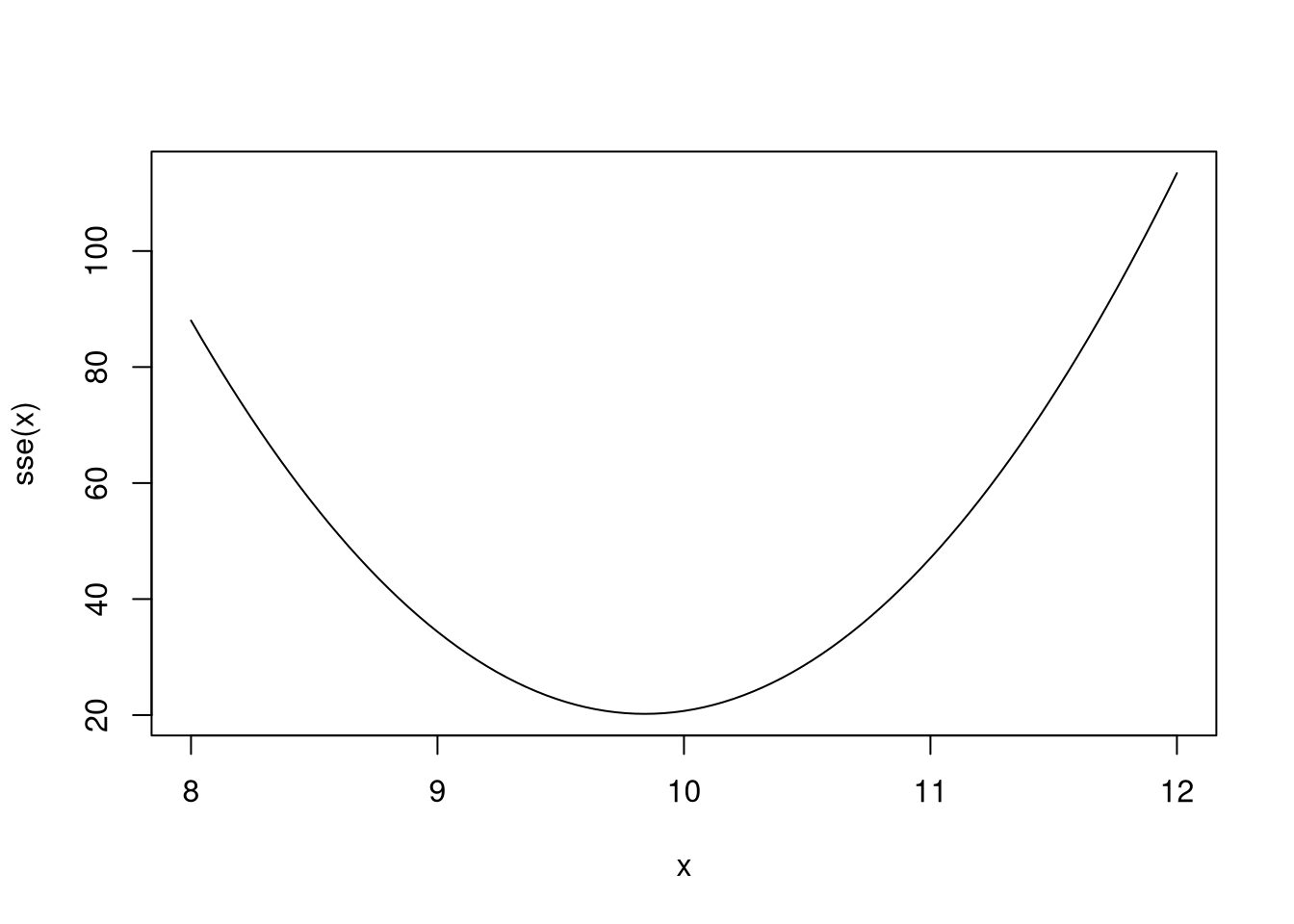
The curve seems to take a minimum near 10 (which is the known mean of the data.) For reference, the mean of the data is 9.8411906. That said, for what value of \(\theta\) is \(SSE(\theta)\) minimized?
For this we can use the function optimize(), which is a numerical optimizer.
The first argument to optimize() is a function to minimize. (If we wish to
maximize a function, recall that maximizing \(f\) is the same thing as minimizing
\(-f\). For this reason, optimization literature generally discusses only
minimization problems since such problems automatically include maximization.)
The second argument is a vector defining the interval over which the function
should be minimized. The function then returns a list with elements minimum
and objective, with minimum being where the minimum is attained (in our
case, the optimal \(\theta\)) and objective the value of the function at the
minimum (here, \(SSE(\theta)\)).
Optimization here is easy:
## $minimum
## [1] 9.841191
##
## $objective
## [1] 20.21141## [1] 9.841191These are of course only some uses for closures. I have used them to gain control over random number generation or to write functions that make predictions from fitted models. As the course progresses, look out for more uses of closures (especially after discussing linear models).
Replacement Functions
Consider the following code:
## a b c
## 1 2 3How odd is this code? Doesn’t it seem strange that a function call can cause the value of an input to that function to change, simply because of assignment?
Actually, the above code is misleading, for it was not the function names()
that was called, but names<-().
## function (x) .Primitive("names")## function (x, value) .Primitive("names<-")A commandment of functional programming is that code should not have side
effects. So a function call f(x) should not modify the state of variables
outside of f nor should x have its value modified. Following this rule helps
make code more easily understood and reasoned about. However, replacement
functions such as names<-() test this principle; while technically adhereing
to at least the spirit of the rule, it at least seems as if they change the
value of their arguments. (That said, one can still reason easily about the
state of the program after this function is called due to the presence of the
assignment operator to the right of a function call.)
We write replacement functions like we would any other function, with the following restrictions:
- Our function name should be wrapped in backticks (like with infix functions)
and end with
<-. - We must have at least two arguments, which we call
xandvalue, withxcoming beforevalue. They correspond tof(x) <- value. - We may have additional arguments, but any additional arguments must be
between
xandvalue. - The function must return the modified copy of
x.
I present a simple (yet stupid) example below, where the function changes the values of vectors in particular positions:
## [1] 1 100 3 4 5 6 7 8 9 10# With a position argument
`modify<-` <- function(x, pos = 1, value) {
x[pos] <- value
x
}
modify(x) <- 20
modify(x, 2) <- -2
x## [1] 20 -2 3 4 5 6 7 8 9 10